What's in a gene?
What do genes make? With third generation genome sequencing, a lot more than we thought.
When we’re discussing what’s in a gene, we should probably take a step back… what even is a gene? The answer to this question is likely to vary greatly between scientists.
Dawkins suggested that they’re selfish elements, which aggressively exert their evolutionary prowess by outcompeting others; us mere mortals simply vessels for our ultra-replicating DNA-encoded masters.
There is evidence for this everywhere, especially as the role of retrotransposons (jumping bits of DNA descended from viruses) comes more into the light, as well as the study of meiosis. The genetic mechanisms at play before, during and after gametogenesis (when sex cells are made) bear credence to this concept of highly competitive, possibly selfish genes.
Some genes, for example, can prove lethal to others, preventing them from being passed on even during the formation of egg or sperm cells. These “meiotic drive” genes manage to destroy any other sex cells that don’t contain a copy. There is an “arms race” going on between genetic elements and even entire chromosomes well before fertilisation has occurred.
Whether genes can be called selfish when collaboration is key in ensuring the survival of an organism is up for debate (the action of selfish meiotic drive genes just described can lead to infertility, so it also might not be the ultimate survival strategy). Is charity altruistic or egoistic? Are genes selfish or selfless? We’ll leave that to philosophical musing.

"I think this question is very important. Most people would focus on the coding part, from translational start site to stop codon.
Others would prefer to extend the definition from the transcriptional start to transcriptional end sites - in such a case you might include any transcribed and biologically functional gene bodies: protein coding genes, small non-coding RNAs, long non-coding RNAs."
- Wilfried Haerty, Evolutionary Genomics Group Leader, Earlham Institute
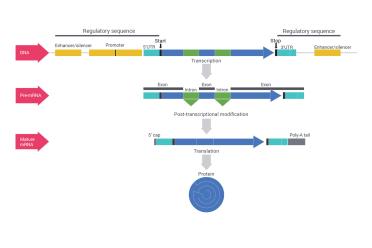
DNA is not just a string of information with genes arranged like beads. These “genes” are composed of many different parts. The "promoter" is where the DNA signals to the correct proteins to come and start reading DNA, while other "regulatory elements" can either turn genes on or off in various ways, or produce products that tune them slightly differently (see our article on miRNA). Within the genes themselves, even when they’re read, there are “introns” which do not end up making any protein but ensure that genes can make different products (proteins, mostly) depending on how they’re cut up and arranged. The DNA essentially is copied to pre-mRNA (transcription), which is then processed to the correct mRNA (via post-transcriptional modification ), which then provides the information necessary to make a protein out of a chain of amino acids (translation).
For now, let’s take a step back to school and explore genes from the widest possible viewpoint. Bits of DNA that encode the proteins that make you you and me me, a squirrel a squirrel and a monkey a monkey.
There are a whole host of reasons why this is over simplistic, such as the now well-documented role of non-coding DNA and RNA, but we’ll go with the protein coding bits for the sake of this article.
Essentially that will do as an explanation, though as our other pieces on “what’s in a genome” and “long non-coding RNAmazing” attest - it’s a bit more complicated than that.
(For the pedantic among us, technically the Central Dogma means that once information is in a protein, you can't get it out again - whatever that means in a world of multiomics).
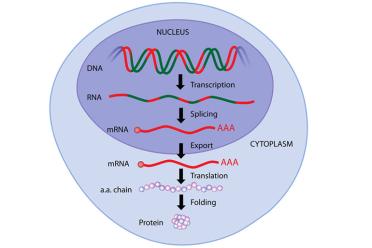
Through several stages, DNA gets “transcribed” to RNA in the nucleus, which becomes mRNA after “splicing”, before being transported to a ribosome where tRNA helps “translate” the mRNA sequence and attaches amino acids in a chain, which then, after “folding” becomes a protein. If none of this makes sense now, don’t worry, carry on reading!
For now let’s imagine that DNA is a long stretch of code, much of which doesn’t really do much, but nearly 2% of which (in a human) is very important indeed.
This 2% contains the “genes” (the protein coding ones at least). These genes are like little instructions that make proteins.
DNA is made of four chemical base pairs that give us a code: A, T, G and C (actual names adenine, thymine, guanine and cytosine). This code makes little three letter words, such as ATG (which starts a gene in most organisms), or TGA (which signals the end of a gene sequence).
These words, also known as codons, are pretty important. They essentially encode which amino acid - the basic building blocks of proteins - should be attached to the next one and in which order. The order that these are attached determines what protein comes out at the end.
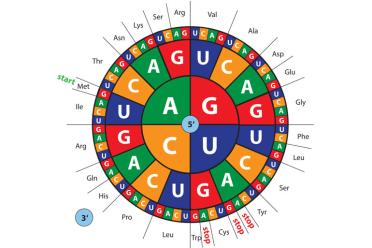
Here we see how the DNA “code” works. In RNA, there is no equivalent of “T” in DNA, instead there is “U” (uracil). The “messenger RNA” (mRNA) relays a message to a “transfer RNA” (tRNA) via three letter words known as codons. Attached to the tRNA is an amino acid - the building blocks of proteins. So, a DNA sequence of GGG ACG CCC would see a glycine next to a threonine next to a proline...
Just one change in a three letter codon can have a significant impact on what amino acid (and therefore what protein) gets attached to the next one… or not.
Indeed, variations on a theme can lead to all sorts of differences, or problems. Cystic fibrosis, for example, is caused by the accidental inclusion of extra nucleotides (the DNA letters A, T, G and C) or a deletion of one or more of them.
Less drastic effects occur even with taste. People who like broccoli or coriander have simple switches in the DNA letters at certain points that set their taste buds apart from those who absolutely detest the taste.

A green fluorescent jellyfish could quickly become blue or red with another simple genetic switch.


The green flourescent protein that makes jellyfish bioluminescent is a very useful scientific tool and can be used to locate where other proteins exist in different life forms.

It’s not simply a case of one gene making one protein, however, unless you’re a bacterium.
In fact, “genes”, or what look like them, are formed in parts - indicated by the “ATG” start codon and the stop signals. In most eukaryotes, these parts are called exons and they are divided by introns.
To make proteins, first the exons are transcribed from DNA into another molecule called “messenger RNA” (mRNA), which is essentially a copy.
In the mRNA copy, the exons are stitched together by removing the introns, and the whole message is then read and translated (via “transfer RNA” at the ribosome) to make a protein, which is essentially a long string of amino acids all joined together and folded in a particular way.
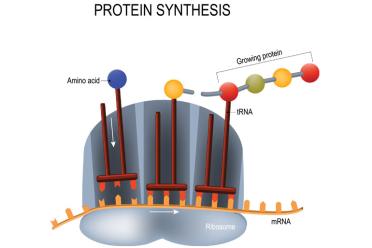
mRNA carries the sequence to be translated at the ribosome by tRNA, which brings along the appropriate amino acids to eventually form a protein.
However, in a gene with 10 exons, not all of them might be included in the final transcripts. It might be that all of them are included, or several are skipped over, leading to totally different messenger RNAs originating from a single gene.
This is called “alternative splicing.”
Splicing is just a fancy term for joining bits of messenger RNA together, but it has far reaching consequences. Depending on which bits get joined together, we get different proteins as a result.
Let’s say we have 10 exons in a gene: let’s call them 1, 2, 3, 4, 5, 6, 7, 8, 9, 10.
The number of permutations from having ten of anything is 10*9*8*7*6*5*4*3*2*1, which is 3628800, which is a lot of permutations.
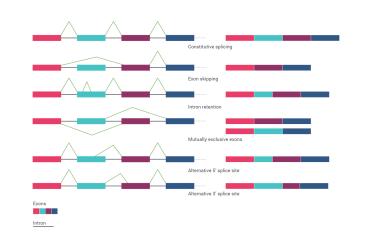
Alternative splicing ensures that one gene does not necessarily make just one protein, but can lead to lots of different possible outcomes. Here, each of the coloured blocks represents an “exon” (the region of a gene that can become protein) and the black bars represent “introns” (the bits of a gene that don’t become protein, but are still very important). Essentially, you can join up the exons in different ways based on the addition of “splice junctions” to each other as you can see from the figure. There are lots of possible combinations.
The actual number of transcripts (messenger RNA copies) from any one gene is nowhere near that - but the more we find out about our genome with ever more sophisticated methods - the more versions of a “gene” we are revealing, which has untold consequences when it comes to novel concepts such as genome editing.
Drosophila melanogaster - the fruit fly - is a popular model system for genetics which has 13, 931 protein coding genes. One of these, Dscam, has 38,016 predicted protein coding transcripts due to alternative splicing.
It’s not just changes in the coding sections of DNA, the exons, that can cause unwanted changes. Sometimes, even a change in one of the non-coding regions, the introns, can have a big effect - especially when it comes down to splicing.

What happens when genes are joined together in the wrong way?

Duchenne myodystrophy, retinitis pigmentosa, leukemia - to name but a few. There are currently investigations underway into whether we can genetically alter DNA via splicing to treat conditions such as Duchenne myodystrophy and Cystic Fibrosis.
One area where aberrant splicing could have unwanted side effects is in the brain, which is a place where small changes can have big consequences. One of these undesired results is schizophrenia, which may be linked to changes in a certain calcium channel encoded by the CACNA1C gene, which may well be an interesting, druggable target.
Why can’t we just edit this section of DNA out and possibly prevent schizophrenia? Here we need to bring in another level of complexity.
There’s a good reason why a gene gives rise to many versions of a similar product, which makes sense in the arena of multicellular organisms (moss, wheat, fruit flies, worms, fishes, mice, humans). The human body contains somewhere in the region of 40 trillion (or so) cells, which form at least 200 different cell types.
The DNA in each and every one of these cells (apart from sperm and eggs) is essentially the same - but each cell looks and behaves differently. That’s because genes are expressed differently in different cells. Some genes are more active, some less active - and there are many degrees of regulation.
One of these regulatory mechanisms is alternative splicing. So, the CACNA1C gene exists in the heart and the brain, but different isoforms exist in either organ, thanks to all of the different exons within the gene that can give rise to similar proteins with different functions.
The problem with this in the era of genome editing is that, while removing schizophrenia, a change in DNA sequence in the CACNA1C gene might also cause the heart to malfunction.
Follow your heart, or your brain? Perhaps both..
This is where recent research at Earlham Institute and the University of Oxford comes in.
They say the human genome has been “sequenced to death”, but further analysis suggests that maybe we ought to go back and take a shrewder look. While it may be true for DNA, leading to a nearly complete genome, with RNA we still have much to discover.
The recent research aimed to take a deeper look at the CACNA1C gene using a new type of long read genome sequencing (namely the Oxford Nanopore MinION), which along with bioinformatics analysis revealed something surprising.
Initial annotations of the genes reported a little over 50 exons and about 40 transcripts. In this gene alone the team discovered an extra 38 novel exons that could give rise to another 83 novel versions of the protein.
In the fruit fly example we mentioned earlier, long read sequencing has already validated 18,000 of the possible 38,016 protein coding transcripts from just a single protein coding gene.

This is both good and bad news - especially when it comes to genome editing for treatment of disease.
On the one hand, it is possible that we can better understand all of the different isoforms of CACNA1C and where they appear, so that we can perhaps target only those versions that appear linked to schizophrenia in the brain while the versions expressed in the heart remain unaffected.
On the other hand, it is clear that we still have much to discover when it comes to alternative splicing, and that editing genes may have umpteen unknown consequences we have yet to discover.

So why is it we have missed so much until now?

The reason is that most modern DNA sequencing methods have relied on “short reads”. When the average human messenger RNA (mRNA) is 2200 letters long, and the average read on the most popular current platform is between 250-800 letters, information can easily be lost - especially when it’s down to computers to piece that information back together.
By sequencing entire sections of mRNA - essentially entire copies of a gene - using technology such as the Oxford Nanopore MinION, we are far less likely to miss information. By taking samples from different tissues, using the long read sequencing, we can truly capture the variation in sequence between tissues in different developmental stages - and reveal far more complexity in the process.
Not just between tissues, in fact, but between cells within tissues in an individual over time - a multidimensional transcriptome, if you will.
We should take care, in an era when editing the genomes of babies is but a step away, to ensure that we really know what we think we know when it comes to genes. Of course, the more information we glean from the human genome using techniques such as those described here - the more we can be sure that we’re progressing down the correct route.
Editing out a genetic ailment is one thing - arguably the best course of action in the face of suddenly losing a child - but clearly altering genes can have unseen consequences. If one “gene” can encode scores of potential proteins, then the simple switch of a codon might not be the simple approach it might have initially seemed.
What is for sure is that with the latest genome sequencing and supercomputing methods available, we are unravelling the complexities of life in greater detail and depth than ever before, which can only improve how we understand and tackle important diseases in the future.