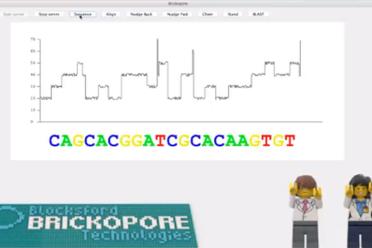
EI also presented its LEGO® DNA sequencer – the Blocksford Brickopore – to the public for the first time. With the recent announcement that Illumina will acquire Pacific Biosciences, Iain Macaulay and Richard Leggett’s creation is a timely addition to a market in need of more competition.
Children, big and small, love LEGO® and its use as a science communication tool has an ever-growing pool of enthusiasts at EI and beyond (check out Monster Lab, for one). The success of EI’s LEGO® sequencer shows why the humble brainchild of a Jutlandian carpenter is so effective. Imagination.
The sequencer is designed to determine the order of red (“T”), green (“A”), yellow (“G”) and blue (“C”) LEGO bricks arranged in a stack of twenty-two with a few white bricks added to one end. In the laboratory, a DNA molecule prepared for real sequencing is referred to as a library and will often have some kind of adapter added on to make it manipulatable.
In this case the white blocks calibrate the sequencer so that it starts to read the sequence at the right position.