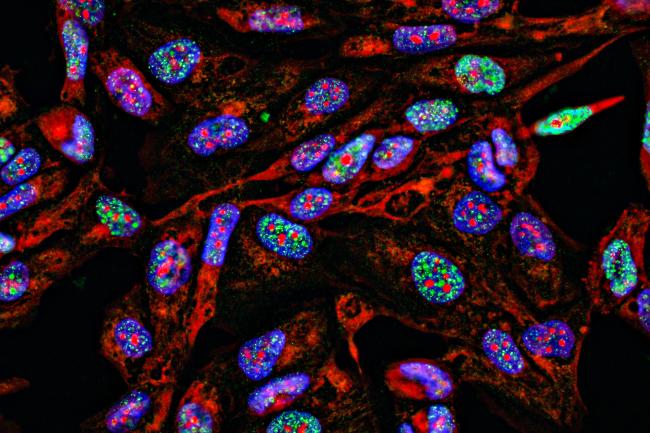
Dr Ellen Cameron, a postdoctoral researcher in the Papatheodorou group at the Earlham Institute, is developing a lichen cell atlas - a comprehensive and detailed map of all cell types in the organism.
“We encounter lichens everywhere in our everyday lives,” she says. “They're globally distributed and ecologically significant, but often overlooked. Right now, I’m using single-cell transcriptomics to explore gene expression in lichens.
“By looking for genes with specialised functions and different expression levels we can begin to disentangle how the fungi and algae interact and form these symbiotic associations. We can also begin to identify the diverse roles individual cells play within the lichen, which is very exciting. ”
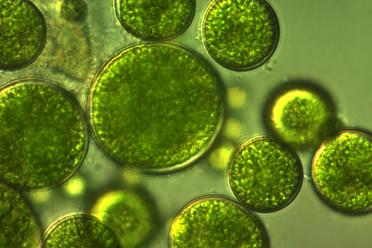
Dr Cameron is working on common orange lichen, a common and widespread brightly coloured lichen growing in very diverse habitats.
Its fungal partner is Xanthoria parietina (the lichen is named after the fungal partner) and the algal partner is a unicellular green algae, a species of Trebouxia.