Using MARTi for taxonomic classification and visualisation of metagenomic data
A step-by-step guide for using MARTi software, from initial setup through to interactive data exploration.
This article is part of our technical series, designed to provide the bioscience community with in-depth knowledge and insight from experts working at the Earlham Institute.
Ned is a Postdoctoral Researcher in the Leggett Group where he leads the development of the front-end for MARTi.
He completed his PhD at the Earlham Institute, where he developed innovative tools and methods for nanopore-based metagenomics and real-time data analysis.
Real-time metagenomics, fuelled by the development of nanopore sequencing - can facilitate a much faster route from sample to results in time-critical situations such as point-of-care diagnostics and bio-surveillance.
Most metagenomic workflows require a sequencing run to complete before analysis can begin, which delays research and slows down diagnostic processes.The sooner you know “who’s there / what genes / what resistances”, the sooner action can be taken.
The full potential of real-time metagenomics remains largely unrealised due to the lack of open-source, offline, real-time analysis tools and pipelines.
MARTi (Metagenomic Analysis in Real Time) is designed to overcome this problem.
MARTi is an open-source software tool that allows users to analyse and visualise metagenomic sequencing data while the sequencing is still in progress. MARTi can also be used once sequencing has finished for less time-critical applications.
Whether you are studying microbial communities in soil, monitoring pathogens in a clinical setting, or exploring the hidden diversity of air samples, MARTi provides insight in real time.
In this blog, you will learn:
how to install MARTi
how to prepare a configuration file
By the end, you will understand how to use MARTi, from initial setup through to interactive data exploration.
Screenshot of the MARTi GUI visualising metagenomic data
The latest version of MARTi is available on GitHub. Detailed installation instructions and user documentation can be found in the MARTi ReadTheDocs pages. Installation is straightforward on Linux and macOS systems.
The easiest way to install MARTi is through mamba, a fast and efficient package manager that works seamlessly with Conda environments. Using mamba ensures that all dependencies are installed correctly and that the software runs in an isolated, reproducible environment.
If you have not already installed MARTi, follow the step-by-step instructions in the installation guide. These instructions cover everything from setting up mamba and creating a new environment to verifying that MARTi has been installed successfully.
For this tutorial, we will also need to install Kraken2 into the same environment:
Installing Kraken2 in the MARTi Environment
# If you created an environment named "marti"
mamba activate marti
# Install Kraken2 into that environment
mamba install -c conda-forge -c bioconda kraken2
# Verify the install
kraken2 --version
Download example data and database
Once MARTi is installed, the next step is to download the data we will use in this tutorial.
We will use four small read sets that have been generated by subsampling from the simulated metagenomic dataset described in the MARTi manuscript. These data represent four mock microbiome communities and provide an ideal test case for learning how to run MARTi. The subsampled datasets are small enough to process quickly on a standard desktop computer but still contain enough diversity to demonstrate key features of MARTi. You can download the data (marti_tutorial.zip) from here.
MARTi can currently classify reads using four different tools: BLAST, DIAMOND, Kraken2, and Centrifuge. Each has its own advantages depending on your computational resources and analysis goals.
In this tutorial we will use Kraken2, a k-mer–based tool, for classification. Kraken2 is considerably faster than alignment-based tools such as BLAST, which makes it well suited to real-time metagenomic analysis and desktop use. For the database, we will use the PlusPF-8 Kraken2 database, which can be downloaded from the Ben Langmead AWS index collection.
We normally recommend BLAST for classification, since it provides highly accurate alignments and well-established scoring methods. However, BLAST is significantly slower and can become computationally intensive when used with large datasets or comprehensive databases such as NCBI’s nt. For BLAST-based classification we recommend running MARTi on a high-performance computing (HPC) system or a powerful workstation to achieve practical runtimes.
After extracting the downloaded file, the next step is to set up the configuration file. This file defines how MARTi processes your data and specifies which tools and databases are used.
The simplest way to set up a configuration file is to generate a template by running the following command, and then edit the file to suit your analysis:
marti -writeconfig config.txt
Run this command to create a default configuration file named config.txt in your marti_example/config directory. Now open this file in your favourite text editor so that we can modify the settings according to our mock microbiome data, reference database, and analysis goals.
First, let’s give the analysis run a name. I’m using “mock_microbiome” for this example. Then, set the RawDataDir path to your unzipped marti_example/reads/mock_communities directory.
Set the SampleDir (Output location for MARTi) to the full path to the new output directory you want MARTi to create in your marti_example/output directory.
As we have used barcodes, we need to tell MARTi which barcodes to process and we can give each barcode a name. Add this barcode information under where you have set the paths. The top section of your config file should now looks something like this:
RunName:mock_microbiome
RawDataDir:/usr/reads/mock_communities
SampleDir:/usr/output/mock_microbiome
ProcessBarcodes:01,02,03,04
BarcodeId1:mix1_original
BarcodeId2:mix2_bact_prev
BarcodeId3:mix3_bif_lacto
BarcodeId4:mix4_pathogen
Set MaxJobs to 1. This is the maximum number of parallel jobs (e.g. BLAST) MARTi will initiate.
MARTi requires NCBI taxonomy files, which provide the hierarchical structure of the NCBI taxonomic tree, to assign reads to the correct taxonomic levels and to apply its Lowest Common Ancestor (LCA) algorithm.
The latest NCBI taxonomy files can be downloaded from the NCBI taxonomy archive. After downloading, extract the archive and provide the path to this directory in your configuration file under the TaxonomyDir setting.
TaxonomyDir:/Path/To/taxdmp_2025-11-01
Remove the BlastProcess section of the configuration and replace it with a Kraken2Process.
Kraken2Process
Name:pluspf-8
Database:/Path/To/Databases/kraken2/k2_pluspf_08_GB_20251015/
options:--confidence 0.01
UseToClassify
Update the “Database” path to point to your extracted k2 database directory.
After you’ve made all these changes you should have a config file that looks something like this (but with your paths).
# MARTi config file
# Lines beginning with # are comment lines.
# The RawDataDir points to the directory containing the fastq_pass directory created by MinKNOW.
# The SampleDir points to the directory where MARTi will write intermediate and final results.
RunName:mock_microbiome_k2
RawDataDir:/Path/To/example_marti/marti_example_blog_tutorial/marti_example/reads/mock_communities
SampleDir:/Path/To/example_marti/marti_example_blog_tutorial/marti_example/output/mock_microbiome_k2
ProcessBarcodes:01,02,03,04
BarcodeId1:mix1_original
BarcodeId2:mix2_bact_prev
BarcodeId3:mix3_bif_lacto
BarcodeId4:mix4_pathogen
# Scheduler tells MARTi how to schedule parallel jobs. Options:
# - local - MARTi will handle running jobs.
# - slurm - jobs are submitted via SLURM (in development).
# MaxJobs specifies the maximum number of parallel jobs (e.g. BLAST) the software will initiate.
Scheduler:local
MaxJobs:1
# InactivityTimeout is the time (in seconds) after which MARTi will stop waiting for new reads.
# It will continue processing all existing reads and exit upon completion.
# StopProcessingAfter tells MARTi to stop processing after X reads have been processed. For indefinite, use 0.
InactivityTimeout:10
StopProcessingAfter:0
# TaxonomyDir specifies the location of the NCBI taxonomy directory. This is a directory containing nodes.dmp and names.dmp files.
TaxonomyDir:/Path/To/Your/taxonomy
# A pre filter ignores reads below quality and length thresholds.
# ReadFilterMinQ specifies the minimum mean read quality to accept a read for analysis.
# ReadFilterMinLength specifies the minimum read length to accept a read for analysis.
ReadFilterMinQ:8
ReadFilterMinLength:150
# ConvertFastQ is needed if using a tool that requires FASTA files (e.g. BLAST).
# ReadsPerBlast sets how many reads are contained within each BLAST chunk.
ConvertFastQ
ReadsPerBlast:4000
# A config file can contain a number of BLAST processes.
# Each BLAST process begins with the BlastProcess keywod.
# Name specifies a name for this process.
# Program specifies the BLAST program to use (e.g. megablast).
# Database specifies the path to the BLAST database, as would be passed to the BLAST program.
# MaxE specifies a maximum E value for hits.
# MaxTargetSeqs specifies a value for BLAST's -max_target_seqs option.
# BlastThreads specifies how many threads to use for each Blast process.
# UseToClassify marks this BLAST process as the process to use for taxonomy assignment (e.g. nt)
Kraken2Process
Name:pluspf-8
Database:/Path/To/Databases/kraken2/k2_pluspf_08_GB_20251015/
options:--confidence 0.01
UseToClassify
# A Lowest Common Ancestor algorithm is used to assign BLAST hits to taxa. Default parameters are:
# LCAMaxHits specifies the maximum number of BLAST hits to inspect in (100).
# LCAScorePercent specifies the percentage of maximum bit score that a hit must achieve to be considered (90).
# LCAMinIdentity specifies the minimum % identity of a hit to be considered for LCA (60)
# LCAMinQueryCoverage specifies the minium % query coverage for a hit to be considered (0)
# LCAMinCombinedScore specifies the minimum combined identity + query coverage for a hit to be considered (0)
# LCAMinLength specifies the minimum length of hit to consider
# LCAMinReadLength specifies the minimum length of read to consider
LCAMaxHits:100
LCAScorePercent:90.0
LCAMinIdentity:60
LCAMinQueryCoverage:0
LCAMinCombinedScore:0
LCAMinLength:150
LCAMinReadLength:0
# Metadata blocks can be used to describe the sample being analysed.
# Each field is optional and can be removed if not required.
# The 'keywords' field is used for searching in the GUI.
Metadata
Location:
Date:
Time:
Temperature:
Humidity:
Keywords:
Tip when using a large database:
When running MARTi with a large database, consider using the “TaxaFilter” parameter in the BlastProcess to exclude certain taxa to minimise wasted computational effort and improve the classification results. For example, the nt database contains entries from the “Other” category (including synthetic constructs, cloning vectors and unclassified environmental sequences).
If a read aligns both to a genuine taxonomic group and to a sequence within “Other,” then MARTi’s lowest common ancestor (LCA) algorithm will assign the read at the root node of the tree, losing any meaningful classification.
By excluding these unwanted taxa upfront you reduce noise, improve the specificity of downstream results and make real-time analysis more efficient.
Once your configuration file is ready you can start MARTi. There are two ways to launch a new analysis: from within the MARTi graphical interface (GUI) or directly from the command line.
In this example, we will run MARTi from the command line. This approach is the same one you would use on a high-performance computing (HPC) system or when running MARTi remotely, and it provides more control over job scheduling and resource use.
To start MARTi from the command line, navigate to your MARTi example directory and run:
marti -config config/config.txtMARTi will begin processing your reads immediately. Progress messages will appear in the terminal as data are analysed. The output directory you defined in your configuration file will be created automatically, containing subdirectories for logs, intermediate files, and final results.
Once the run has started, you can explore the results using the MARTi GUI. The GUI provides an interactive way to view the results generated during the run within a web browser.
Monitoring your run in this way provides valuable insight into how your sample is behaving during sequencing. You can quickly see which taxa are dominant, identify potential contaminants, and assess whether it is worth continuing the sequencing run. This immediate feedback is one of the key advantages of real-time metagenomic analysis with MARTi.
To get started with the GUI, run the following command and provide it with the path to the marti_example/output directory:
marti_gui --marti /path/to/marti_example/outputWhen the GUI starts, it will launch a local web server.
Open a web browser and navigate to http://localhost:3000 to access the interface. From there, you can browse through each barcode or sample, view classifications at different taxonomic levels, and explore visualisations of their taxonomic content.
The Samples page allows you to view all samples detected in the MARTi output directory. Each entry represents either an individual nanopore sequencing run or a barcoded sample within a run. From here, you can choose which samples to load into the Dashboard or Compare analysis modes.
To open a sample in Dashboard mode, simply click on its sample ID or select the dashboard icon next to it. The selected sample will then load with its own set of visualisations.
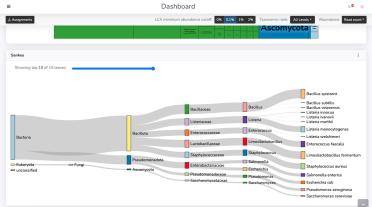
The Dashboard page is where you can explore the analysis results for an individual MARTi sample in detail. A sample may be one that has completed processing, or one currently being analysed by the MARTi Engine. When MARTi is running in real time, the Dashboard updates automatically as new reads are classified, allowing you to monitor changes in community composition as sequencing progresses.
The Dashboard presents a series of interactive plots and tables that summarise the taxonomic content of the sample. In the example video below, we scroll down to the Sankey plot, which shows the flow of reads through the taxonomic hierarchy, highlighting how classifications are distributed across major groups.
Tip: Each visualisation on the Dashboard includes a set of options in the top right-hand corner for customising and exporting the figures. These tools allow you to change the display, filter results, or save publication-ready images directly from the interface.
The Compare page enables you to explore multiple MARTi samples side by side, including samples that are still being analysed in real time.
To use the Compare mode, first return to the Samples page. On the left-hand side, each sample has a tick box that allows you to select it for comparison.
In this tutorial, we will select all four barcoded samples. Once the samples are selected, click Compare to load them into the multi-sample view.
The Compare page provides several visualisation options that summarise taxonomic profiles across samples. In the demonstration below, we scroll down to the heatmap plot, which displays the relative abundance of detected taxa across the selected samples. Heatmaps are particularly effective for highlighting shared taxa, sample-specific organisms, and overall differences in community structure.
This tutorial has introduced the core steps involved in using MARTi for real-time taxonomic classification and visualisation of metagenomic data.
We have explored how to install MARTi, prepare a configuration file, and run an analysis from the command line. We have also demonstrated how the MARTi GUI can be used to monitor analyses in real time, inspect individual samples in detail, and compare multiple samples side by side.
Our aim in developing MARTi is to help researchers make the most of real-time nanopore sequencing by providing an accessible, flexible, and open-source framework for metagenomic analysis.
Real-time insight into community composition can guide experimental decisions, highlight contamination or unexpected organisms, and support faster responses in time-critical settings such as environmental monitoring or pathogen detection.
We hope this walkthrough helps new users gain confidence in setting up and running MARTi with their own data. We plan to build on MARTi’s capabilities by adding additional features, visualisations, and reporting options.
Feedback, suggestions, and contributions from the community are always welcome.
MARTi was developed with support from the Biotechnology and Biological Sciences Research Council (BBSRC) through the Norwich Research Park Biosciences Doctoral Training Partnership (NRPDTP), Earlham Institute Decoding Biodiversity Institute Strategic Programme, and cross-institute Delivering Sustainable Wheat programme.
The latest version of the MARTi software is available from Github under the MIT License.
Documentation can be found on Read the docs.io.
Additionally, an installation-free demo of the MARTi GUI is available at Cyverseuk.
If you have any questions or feedback about the content in this article, please contact: communications@earlham.ac.uk.