Focus on fungi helps fight global threat to our food
Fungal attacks on the world’s most important crops are one of the biggest threats to food security, with their impact likely to worsen as temperatures rise and fungi expand their range.
Plant pathogen invasion is a focus of research at the Earlham Institute, with several scientists working on the global fight to tackle fungal pathogens.
Crops under the microscope include wheat, beet, and bananas.
Fungi can travel long distances, remain dormant in soil for years, and are extremely adaptable. These advantages give them everything they need to be a major destroyer of crops.
Dr Rowena Hill is a Postdoctoral Research Scientist in the Neil Hall Group at the Earlham Institute. She is collaborating with Rothamstead Research to investigate the destructive wheat pathogen Gaeumannomyces tritici, also known as wheat take-all.
She explains the pathogen acquired its common name because it can kill entire fields of wheat, taking every plant.
“Take-all is a fungal infection which affects the root,” she says. “With other pathogens, which affect the leaves and stems, it’s often easier to detect their presence, but take-all is not easily detectable without destroying the plant.
“It’s cumulative - the first year it’s in a field there are usually no symptoms. If a farmer continues to grow wheat in that field, the fungus builds up in the soil and, in the following two or three years, it kills all the plants.
“We’re hoping that eventually we’ll be able to develop an assay which will allow farmers to test soil in fields for the presence of the fungus.”

Fungicides are of limited effectiveness against take-all, which primarily affects the UK winter wheat harvest. As it occurs below ground, it’s difficult to determine the levels of damage caused. But estimates suggest the disease affects half of UK wheat crops, and can cause average yield losses of up to 20 per cent.
A soil test would be a game-changer for farmers, allowing them to rotate crops out of the fields before take-all strikes. It’s one of the ambitions of Delivering Sustainable Wheat (DSW) - a collaborative strategic research programme led by the John Innes Centre.
As part of DSW, Rothamsted Research is providing experimental data from wheat fields, and the Earlham Institute is sequencing the genome and analysing bioinformatics.

It’s cumulative - the first year it’s in a field there are usually no symptoms. If a farmer continues to grow wheat in that field, the fungus builds up in the soil and, in the following two or three years, it kills all the plants.

Dr Rowena Hill
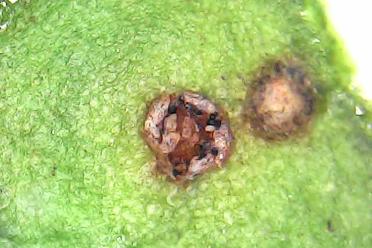


Pictured from L-R: 1. Uromyces beticola on sugar beet leaf (1000x magnification). ©Michelle Grey/Earlham Institute. 2. Spots of disease showing on a beet leaf. 3. Beta vulgaris subsp. maritima, Sea Beet, is the close relative of Sugar Beet.
Rust fungi, a common family of pathogens, have a huge impact on crops worldwide. They particularly affect cereals.
CASE PhD student Hélène Yvanne, also in the Hall Group, is working with German company KWS to detect a resistance gene to Uromyces beticola, or beet rust. Yvanne is using beet rust as a model to understand how wild crop relatives could be used as a source of resistance.
Sugar beet is closely related to sea beet, which grows wild around coasts all over Europe, and the two share the fungal pathogen. Yvanne is comparing the genomes of more than 500 sequenced beets.
“Sugar beet was domesticated recently and has not diverged far from sea beet,” says Yvanne. “This means it’s relatively easy to see where the genomes diverge.
“Since they share the same pathogens, this gives us an opportunity to study the evolution of pathogens between sugar beet crops and wild sea beet and potentially identify the genes affecting susceptibility and resistance.”
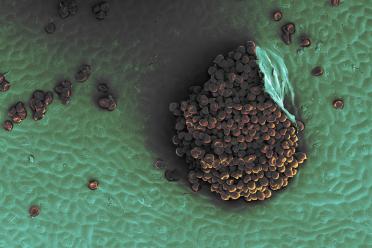
Insight into the evolution of pathogens and resistance in wild crop relatives is specifically useful for crops.
Dr Mark McMullan, Population Genomicist in the Hall Group, works with both Hill and Yvanne. He is using signals of adaptation in the genes of rusts infecting wild and crop beets to identify evolving genes which could be used in agriculture.
The concept of understanding pathogen evolution in this way came from work on the fungal pathogen causing ash dieback, a disease of ash trees which kills approximately 70 to 85 per cent of the trees it infects.
This pathogen evolved in Asia on Manchurian and Chinese ash trees, which have resistance, and it doesn’t cause much damage to them. But European ash has evolved no defences and the pathogen’s introduction to the continent was devastating.
McMullan is applying bioinformatics and population biology to understand the evolution of genes important for pathogen virulence and host resistance.
The long-term implication of this work is in developing surveillance strategies for invasive pathogens, as well as resistant hosts.

This gives us an opportunity to study the evolution of pathogens between sugar beet crops and wild sea beet and potentially identify the genes affecting susceptibility and resistance.

Hélène Yvanne
Elsewhere in the Earlham Institute, Carolina Olave Achury, based in Jose De Vega’s group, is studying resistance to Panama disease - a fungus which is infecting and destroying the world’s banana crops.
Around half of all bananas grown worldwide are of the Cavendish variety, accounting for the overwhelming majority eaten in the western world. But these plants are all genetic clones, making the Cavendish incredibly vulnerable to the emergence of new threats.
Olave Achury’s research is using genomics and data science to find sources of resistance in other banana cultivars.
Thousands of banana varieties are cultivated in the tropics of Asia and one of these may hold genes that offer resistance to the pathogen, which they hope can be quickly introduced to the Cavendish plants.

Michelle Grey, Research Assistant in the Hall Group, has done wet-lab work for all three of the Hall Group projects and has a strong interest in plant pathogens.
“I have been fascinated by how plants respond to pathogens since I was at school,” she says. “Looking at how the pathogen breaches the plant’s defences and how the plant responds is very interesting to me."

“Now we have genomics, so we can analyse what is actually happening at the genetic level - the resistance genes, the genes which are switched on and why, and what happens when a new pathway opens. We can begin to analyse exactly what the fungus is doing within the plant.
“Being able to engineer resistance into a crop would be a win-win. It would cost farmers less - treating fields for fungi is enormously expensive and has to be repeated when they adapt to it.
“It would also stop run-off from fungicides, which would benefit the environment.”

This knowledge will also uncover some of the mechanisms behind virulence and resistance, and identify new resistance genes that can be deployed in agricultural systems.

Prof Neil Hall
The Earlham Institute is one of many organisations working on plant pathogens, thanks in large part to funding from the Biotechnology and Biological Sciences Research Council, part of UKRI.
Protecting the world's crops from fungal disease, however, requires a unified approach - bringing together farmers, the agricultural industry, plant breeders, biologists, governments, policymakers, and funders.
Professor Neil Hall, Director of the Earlham Institute, recognises the urgency and potential value of the different projects being delivered by his and other groups.
“Nature already holds many of the solutions that are desperately needed to develop sustainable agricultural systems,” he says. “The plant pathogen work at the Earlham Institute uses our expertise in data-driven science to uncover genomic diversity in pathogens and hosts.
“This work helps us understand what genes have been selected for resistance, and how pathogens evolve in response. This knowledge will also uncover some of the mechanisms behind virulence and resistance, and identify new resistance genes that can be deployed in agricultural systems.”