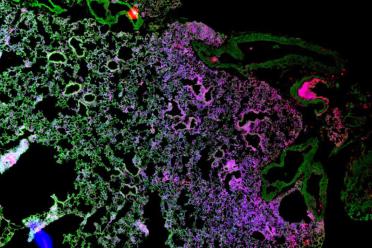
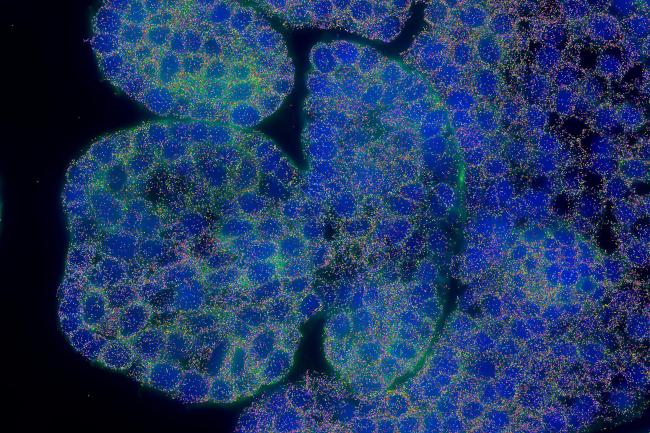
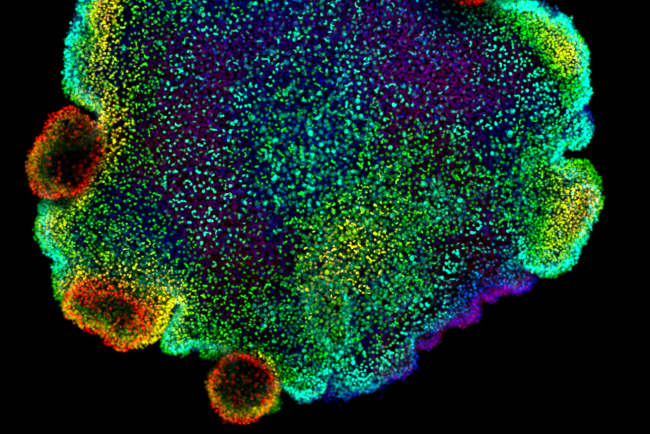
At the Earlham Institute, we combine our computational expertise in single-cell genomics with cutting-edge data science and AI to work on global cell atlas projects.
Scientists in the Papatheodorou Group work across the pipeline of atlas curation, including metadata management and study integration, to data analysis,visualisation, and inference.
We’re collaborating widely with partners in the Biodiversity Cell Atlas, Human Cell Atlas, in particular atlases for Gut and Crohn’s disease, as well as comparing species (human, plant, mouse, fruit fly, lichen) to identify cell-type-specific gene expression patterns.
Cross-species analysis allows scientists to map and compare how cell function and gene expression differ across evolutionary distances, helping us better understand the relationship between gene expression, phenotype and function. This insight will aid global initiatives to conserve biodiversity by increasing our understanding of different species vulnerabilities given environmental stressors.
The Group is also contributing to the Plant Cell Atlas (PCA) initiative, which is comprehensively describing plant cell types and their spatial organisation using high-resolution data, by developing our own single-cell sequencing and analysis platforms.
The compiled atlases can help to answer complex biological questions about cellular function, diversity, and evolution - how cells differ across species, and understanding the composition of tissue within an organism.
In the long-term cell atlas analysis could help with the development of biomarkers for different diseases, repurposing existing drugs for multiple diseases. In agriculture it could enable a better understanding of the complex interactions between pests and the plants in order to develop targeted treatments.
“It’s really exciting to see the new experimental methods being coupled with developments in machine learning and AI, and how they are really increasing our insights in such a rapid way,” Irene adds.