Metagenomics is the study of all organisms present in a particular environment, such as soil, water, or the human body. A key part of metagenomic analysis is understanding what species are present (classification), how much of each there is (abundance), and the function of the microorganisms present.
Real-time metagenomics - the immediate analysis of data while sequencing is in progress - holds the potential to speed up the detection, monitoring, and response to microbial threats in a multitude of settings, including agricultural, environmental, and biosecurity.
However, one of the key barriers to realising the full potential of real-time metagenomics is the lack of flexibility in current tools available, increasing the time it takes for sample analysis to be returned to the site of application.
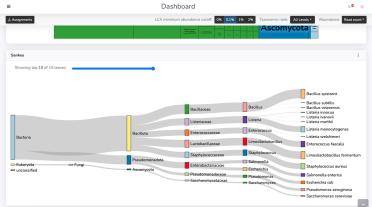
MARTi (Metagenomic Analysis in Real Time), published today in Genome Research, is an open-source software tool that powers real-time analysis and visualisation of metagenomic data. The team have created an accessible interface which increases the usability and accessibility of metagenomic analysis.
Dr Ned Peel, first-author and postdoctoral research scientist at the Earlham Institute said: “It’s operationally very lightweight, you can use it in-field for taxonomic classification on a standard laptop, or undertake larger, complex analysis using high-performance computing.”
“One of the key impacts of MARTi is the immediacy of analysis results - it could be very useful in situations where a quick identification is essential.” added Ned.
In clinical environments this could allow clinicians to rapidly speed up the identification and targeted treatments of pathogen infection.