Stay at home, protect the NHS, save lives. Wash your hands for 20 seconds and avoid touching your face. Keep two metres apart at all times. While the public health advice to combat COVID-19 has been undeniably simple, the virus itself remains an enigma.
The global death toll is well in excess of 250,000, with some patients fighting for their lives in intensive care units, yet others seemingly shrug it off with no symptoms to suggest they were ever infected.
Since the SARS-CoV-2 virus began spreading rapidly around the world, scientists have been racing to understand its pathogenicity. Global collaborations and open data sharing have yielded vast amounts of data which the bioscience community is using to find a potential treatment to reduce the mortality associated with COVID-19.
The mounting evidence points to a peculiar feature of SARS-CoV-2. Far from being limited to the respiratory tract, its deadly effects have been shown to act all over the body - from the lungs to the gut and the kidneys - even causing strokes in otherwise healthy, younger patients.
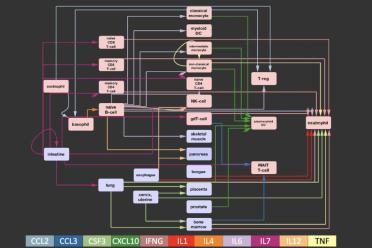
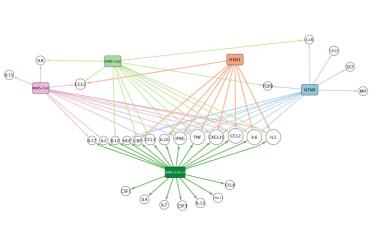
One particularly dangerous effect, which occurs in some patients between 7 and 10 days after infection, is a cytokine storm. The virus essentially causes the immune system to go into overdrive. Struggling to cope, the body can fill the lungs with fluid, cause multiple organ failure, and perhaps even lead to some neurological effect.
If we are to stand any chance of developing effective treatments - and managing this pandemic - we have to find out what is driving the response of our body to SARS-CoV-2.