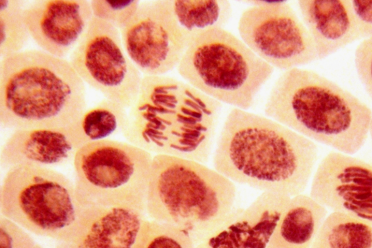
We are trying to understand how blood stem cells differentiate. What mechanisms are driving the differentiation process, and are some stem cells more primed to become a specific type of mature blood cell? Are they more prone, early on, to become one of the different cell types at the end?
I'm looking at the effect of ageing - what happens to these stem cells in the blood of an older organism.
Stem cells have the unique abilities of generating their daughter stem cells (self-renewal) as well as mature cell types (differentiation). Mature blood cells are predominantly short lived and adult hematopoietic stem cells guarantee replenishment of the mature cells during an organism’s lifespan. The delicate balance between self-renewal and differentiation properly regulates stem cell number and guarantees tissues homeostasis, function and repair. When we age this delicate balance seems to be impaired.
It's quite well known that with ageing there is a loss of function of hematopoietic stem cells, but they increase in number. So, we have more, they seem activated, we see an expansion of the stem cell, yet they lose their ability to produce a balance of multiple types of blood cells. We don't really know the mechanism driving that.
We also don't know how different stem cells actually age differently, or whether there’s a shared mechanism between all of them, and how this translates into loss of function. That's one of the questions I’m trying to answer.