The research shows a single letter change in their DNA code can potentially decide whether a plant is a lark or a night owl. The findings may help farmers and crop breeders to select plants with clocks that are best suited to their location, helping to boost yield and even the ability to withstand climate change.
The circadian clock is the molecular metronome which guides organisms through day and night - cockadoodledooing the arrival of morning and drawing the curtains closed at night. In plants, it regulates a wide range of processes, from priming photosynthesis at dawn through to regulating flowering time.
These rhythmic patterns can vary depending on geography, latitude, climate and seasons - with plant clocks having to adapt to cope best with the local conditions.
Researchers at the Earlham Institute and John Innes Centre in Norwich wanted to better understand how much circadian variation exists naturally, with the ultimate goal of breeding crops that are more resilient to local changes in the environment - a pressing threat with climate change.
To investigate the genetic basis of these local differences, the team examined varying circadian rhythms in Swedish Arabidopsis plants to identify and validate genes linked to the changing tick of the clock.
Dr Hannah Rees, a postdoctoral researcher at the Earlham Institute and author of the paper, said: “A plant’s overall health is heavily influenced by how closely its circadian clock is synchronised to the length of each day and the passing of seasons. An accurate body clock can give it an edge over competitors, predators and pathogens.
“We were interested to see how plant circadian clocks would be affected in Sweden; a country that experiences extreme variations in daylight hours and climate. Understanding the genetics behind body clock variation and adaptation could help us breed more climate-resilient crops in other regions.”
The team studied the genes in 191 different varieties of Arabidopsis obtained from across the whole of Sweden. They were looking for tiny differences in genes between these plants which might explain the differences in circadian function.
Their analysis revealed that a single DNA base-pair change in a specific gene - COR28 - was more likely to be found in plants that flowered late and had a longer period length. COR28 is a known coordinator of flowering time, freezing tolerance and the circadian clock; all of which may influence local adaptation in Sweden.
"It’s amazing that just one base-pair change within the sequence of a single gene can influence how quickly the clock ticks," explained Dr Rees.
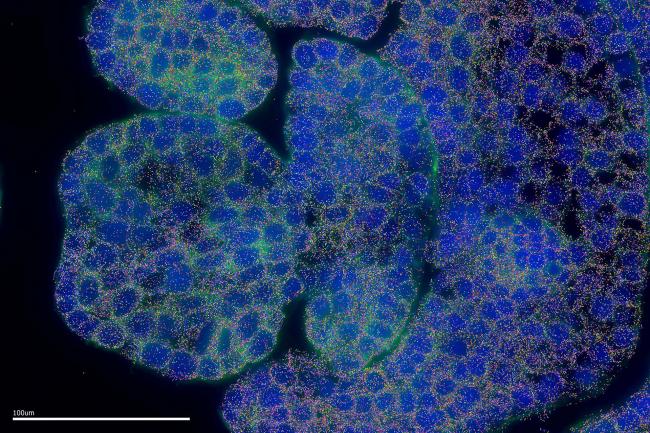
The scientists also used a pioneering delayed fluorescence imaging method to screen plants with differently-tuned circadian clocks. They showed there was over 10 hours difference between the clocks of the earliest risers and latest phased plants - akin to the plants working opposite shift patterns. Both geography and the genetic ancestry of the plant appeared to have an influence.
“Arabidopsis thaliana is a model plant system,” said Dr Rees. “It was the first plant to have its genome sequenced and it’s been extensively studied in circadian biology, but this is the first time anyone has performed this type of association study to find the genes responsible for different clock types.
“Our findings highlight some interesting genes that might present targets for crop breeders, and provide a platform for future research. Our delayed fluorescence imaging system can be used on any green photosynthetic material, making it applicable to a wide range of plants. The next step will be to apply these findings to key agricultural crops, including brassicas and wheat.”
The results of the study have been published in the journal Plant, Cell and Environment.