Biography
I am a Senior Research Assistant in the Technical Genomics Group.
I joined the Earlham Institute in 2015; initially working on the high throughput BAC sequencing project contributing to the reference genomes of important crops such as bread Wheat, Barley and Lolium and gaining extensive experience in sequencing operations using our suite of Illumina platforms.
I subsequently moved into the Library Construction Team and gained a breadth of experience in performing quality control on incoming samples as well as performing on bench and high throughput library construction on a range of sample types using our range of automation platforms.
During the COVID-19 pandemic, EI set up a COVID testing lab and I was part of the team responsible for preparation of reagent plates for the various workstations in the testing lab.
My current role involves running established protocols on samples to perform sample quality control, library construction and sequencing as well as being involved in the development and optimisation of new technical workflows within the group.
Publications
Related reading.

From idea to innovation: inspiring entrepreneurship at Earlham Institute
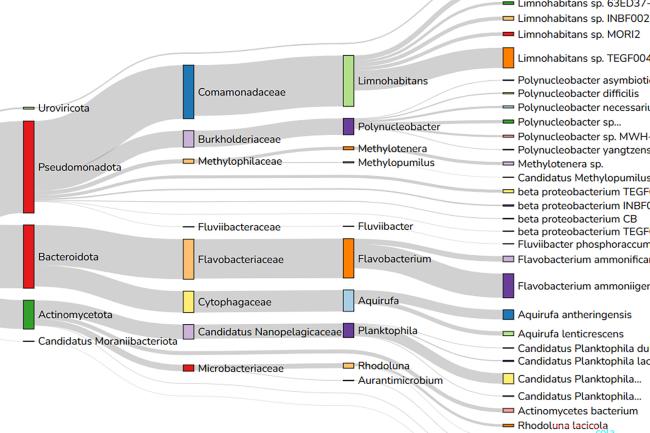
Pangenome annotation opens up a multiverse of genes

Every cell tells a story: single-cell analysis in forensic science

AI and life sciences: why FAIR data is essential

Analysing cell imaging data at scale with AI
LITE2 takes affordable genome sequencing to a new level

Mapping cell diversity with long-read sequencing and single-cell genomics

Earlham Institute and Natural History Museum launch deep tech startup Agnos Biosciences™

New software tool MARTi fast-tracks identification and response to microbial threats.

New BBSRC funding supports expansion of transformative spatial science

Director appointed to lead transformative digital research infrastructure initiative

Devastating crop pathogens can be found by sequencing the air

UKRI given green light for game-changing BioFAIR investment

Earlham Institute begins testing air across Norfolk for a year

Earlham Institute spinout TraitSeq to transform agricultural sector