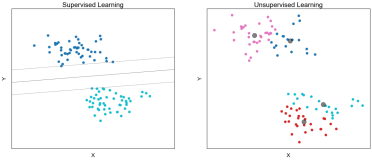
Clustering: An unsupervised method of grouping observations based on similarities in the values of their features.
Data wrangling: The process of restructuring, cleaning or organising a dataset in order to facilitate analysis or visualisation.
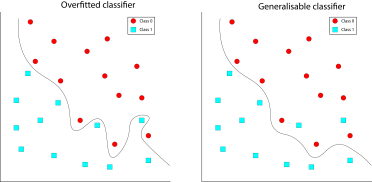
Deep architecture: A neural network model that has multiple intermediate layers. The more layers that the model has, the more complex the patterns it is able to utilise within the dataset.
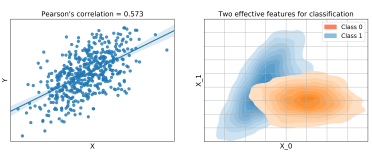
Feature: An explanatory variable that has been recorded as part of a dataset.
Feature engineering: Extracting and generating features from raw data to quantify the observed patterns in a more model friendly way, leading to better results.
Fitting: The procedure of adjusting a model’s weights to more accurately map input to output. Gradient descent methods are most commonly used to calculate how a model’s weights should be changed throughout the fitting (training) procedure.
High dimensional data: A dataset where each observation possesses a high number of features.
Hyperparameters: Parameters of the model that affect the fitting of the model. They can be highly influential on the model’s performance and are typically related to the complexity of the model or the speed and duration of the fitting procedure.
Target: The property/characteristic/variable that we are trying to predict in a supervised machine learning task, sometimes referred to as the label or ground truth.
Weights: Trainable and non-trainable parameters in the model that quantify how the features are mapped from input to output, or input to the next layer in a neural network.