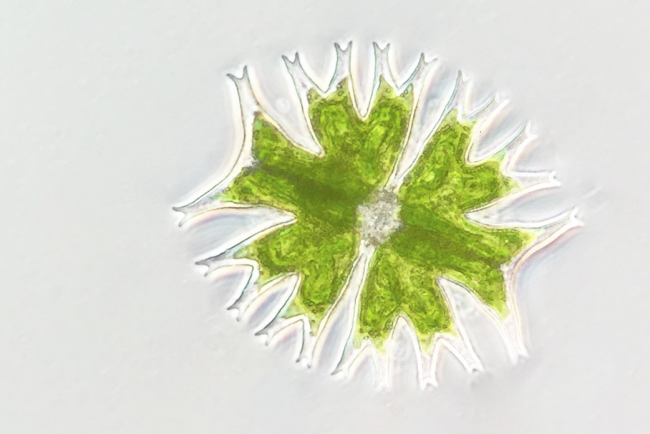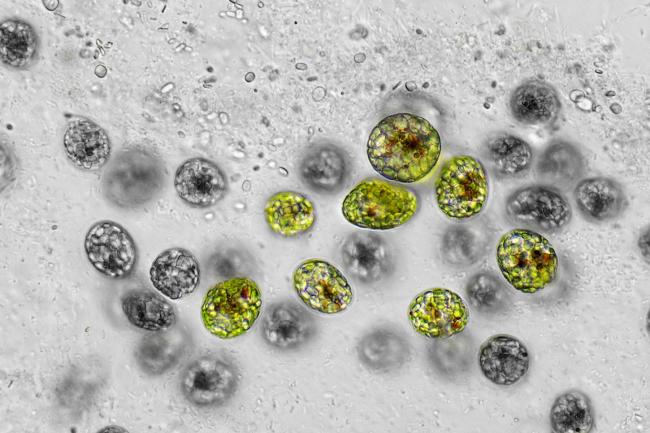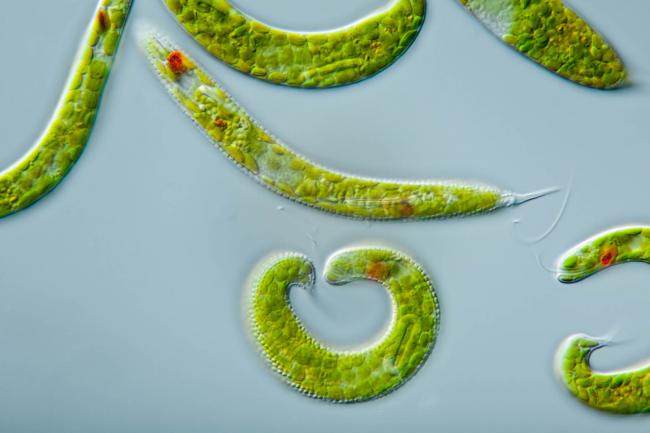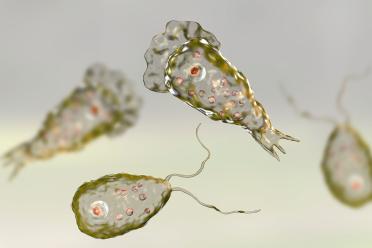
Researchers from the Earlham Institute sequenced the genomes of two uncultivated Naegleria species, collected from the River Leam in Royal Leamington Spa.
Naegleria are found in soils and freshwater worldwide. Of the 47 recognised species, only one - Naegleria fowleri - is pathogenic to humans. It causes primary amoebic meningoencephalitis (PAM) - a rare but almost always fatal brain infection.
While other members of the Naegleria genus aren’t known to directly infect humans, they can still pose a threat to human health, by acting as hosts for bacterial pathogens. These can live inside the amoeba, using it as a shield to persist and multiply in the environment.
The newly sequenced species are close relatives of N. fowleri. And researchers were able to generate two highly complete Naegleria genomes and two complete bacterial genomes belonging to the Legionellaceae family, from just two individual cells.
“This is part of our work with the Darwin Tree of Life project, using single-cell genomics to sequence protists from the environment,” said Dr Jamie McGowan, who led the work at the Earlham Institute, in collaboration with Professor Tom Richards’ group at the University of Oxford.
This discovery was made possible through McGowan’s collaboration with the Earlham Institute’s Technical Genomics Group where Senior Research Assistant James Lipscombe adapted existing protocols to sequence single cells from environmental samples.
Close relatives
The pair are closely related to the brain-eating amoeba, but these cousins of the pathogen aren’t dangerous.
“After analysing the two new genome sequences, we learned they are sister species of the brain-eating amoeba - Naegleria fowleri,” said Dr McGowan, who is now working at University College Dublin.
“Hopefully, by sequencing relatives, we can learn more about the pathogenicity of the brain-eating amoeba. Studying non-pathogenic relatives could give us insight into how the pathogen functions.”
Both amoebae were found to be harbouring novel Legionella bacteria. One genome was closely related to Legionella lytica - a known Naegleria pathogen - while the other represented an early-diverging lineage within the family.
“Coincidentally, when we sequenced these we discovered that they both have a bacterium associated with them—they’re both harbouring Legionella,” McGowan said.
“Legionella is another really nasty infection. It can infect a wide range of hosts: humans, but also very diverse protists. That’s how these bacteria survive in the environment, by living inside protist hosts.”
He says it’s difficult to draw conclusions about the nature of the host-pathogen interaction from just two cells.
“Without more information it’s difficult to say if the amoebae have been infected with Legionella or if the bacteria have a symbiotic relationship with them,” he says.
“Protists can have bacteria associated in different ways - they eat them; bacteria can live on the outside of the cell; or they could be infected."
Single-cell showcase
This work shows the advantages of single-cell sequencing when it comes to disentangling complex genetic relationships. The insights generated were achieved thanks to the unique blend of specialist expertise and infrastructure in single-cell genomics and annotation within the National Bioscience Research Infrastructure in Transformative Genomics.
“Through single-cell genomics, we were able to sequence both the amoeba genome and the bacterial genome,” McGowan explained.
“If we had used something like metagenomics, we wouldn’t be able to link the two. But with single-cell genomics we can tease out the bacterial genome from the protist genome, while linking the bacteria to its protist host. It’s a really powerful technique to understand host-pathogen interactions and how they evolve over time.”
Dr McGowan believes scaling up the approach could reveal much more about the ecology and evolution of Naegleria and Legionella, and microbe-microbe interactions in general.
“Here we just captured two, and we learnt a lot from them,” he said. “If we went out and specifically looked for these organisms and sequenced a large number, we could begin to work out the relationships between the host and pathogen and how they evolve over time.”
The findings spotlight the promise of single-cell genomics for illuminating the hidden worlds of microbial interaction - worlds that may ultimately shape the emergence of new human pathogens.