New research published today in PLOS ONE reveals an improved annotation of microRNAs in the dog genome to further understand its biological role. Providing a platform for future studies into biomedicine, evolution and the domestication of important animals including dogs, cows, horses and pigs.
MicroRNAs (miRNAs) are small non-coding RNA molecules that play a crucial role in regulating gene expression in animals and plants. Using the latest dog genome assembly and small RNA sequences of nine different dog tissues including skin, blood, ovaries and testes, scientists from EI have identified 91 novel miRNAs.
This study provides a significant opportunity not only to enhance our understanding of how miRNAs regulate a variety of biological processes in an important model species for studying human diseases but can lead to further, similar research into the role that miRNAs play in animal domestication.
Lead researcher, Dr Luca Penso Dolfin from EI’s Regulatory & Environmental Genomics Group (Di Palma Group), said: “As miRNAs are so important in orchestrating a variety of cellular processes, the discovery of these 91 novel miRNAs provides a vital starting point to explore their potentially major effects on gene regulation.”
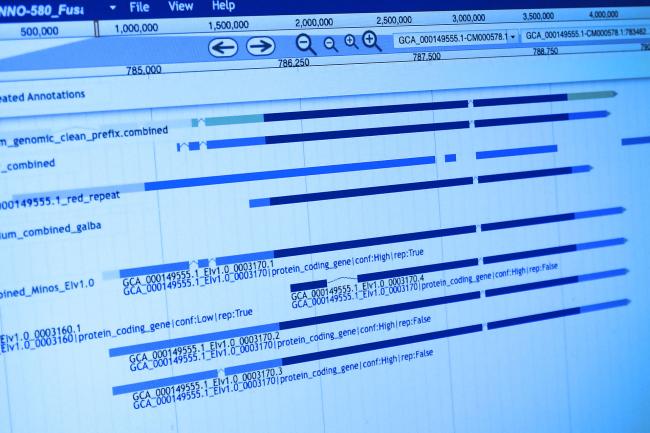
Overall, 811 miRNAs were analysed by Dr Penso-Dolfin: 91 novel microRNA sequences and 720 conserved (that is, common to other organisms). Among these conserved loci, 207 had not previously been identified as canine microRNAs.
Dr Penso-Dolfin, added: “Our results represent a clear improvement in our knowledge of the dog genome, paving the way for further research on the evolution of gene regulation, and the contribution of microRNAs to pathological conditions. We are now looking at additional data for dog and a variety of farm animals, combining microRNA discovery to the investigation of their possible role in domestication.”
The domestic dog, Canis familiaris, is the result of wolf (Canis lupus) domestication, which started around 10,000 years ago. Since then, hundreds of dog breeds have been artificially selected, leading to very high levels of morphological and behavioural variation. Having shared the environment with humans ever since its appearance, the dog has been exposed to similar pathogens and therefore represents an important model system for the study of human diseases.
The publication of the latest Canine genome build and annotation, CanFam3.1 provides an opportunity to enhance our understanding of gene regulation across tissues in the dog model system.
This study was led by EI in collaboration with the Broad Institute, US, and Uppsala University, Sweden. The paper titled: “An improved microRNA annotation of the canine genome” is published in PLOS ONE. Professor Federica Di Palma, Director of Science at EI and Dr Simon Moxon, Project Leader in the Regulatory Genomics Group at EI are joint corresponding authors.
EI is strategically funded by BBSRC and operates a National Capability to promote the application of genomics and bioinformatics to advance bioscience research and innovation.
Image: Gary Kramer