New in 2022: The International Year of Aquaculture and COP15
Recent history confirms how tough it is to make wholly accurate predictions, but we reckon 2022 is set to be a big year for researchers at the Earlham Institute.
Recent history confirms how tough it is to make wholly accurate predictions, but we reckon 2022 is set to be a big year for researchers at the Earlham Institute.
From exciting new, expert-led training workshops to decoding biodiversity and engineering biological solutions to global challenges, we look ahead at what the new year will bring.
While 2022 will be the year of the tiger for many, for us here at the Earlham Institute it’s the year of the fish. Specifically, tilapia.
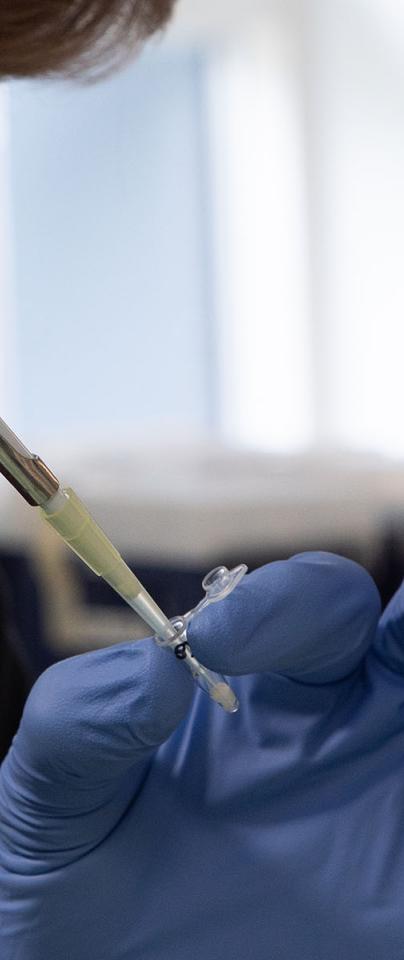
“Small in scale, big in value” - the FAO marks this the International Year of Artisanal Fisheries and Aquaculture, a topic at the heart of our research. Along with WorldFish and collaborators at the Roslin Institute, we’re planning to make quite a splash this year with a brand new genome and annotation for GIFT tilapia, one of the most popular freshwater-farmed fish on Earth.
The research, along with other developments in the Evolutionary Genomics Group - such as tools to spot hybrid fish and insights into cichlid evolution - will give breeders the tools they need to help smallholder farmers get the most out of their ponds, improving socio-economic wellbeing and global food security.
Image below supplied by WorldFish

Inextricably tied to the climate crisis, biodiversity loss is so rapid that humans currently compete with meteors and supervolcanoes in the drivers-of-mass-extinction stakes.
If we’re to prosper, we must collectively cure this current malaise so that we might conserve and continue to benefit from biodiversity. The global community has an opportunity to forge the policies that might begin to make amends this year at the Convention on Biological Diversity (COP15) later this year in Kunming, China.
While the change we need must happen at the political level if we’re to make inroads, the global science community is helping to enable a transition to a world that protects and understands biodiversity for future generations.
At the Earlham Institute, we’ll have the results of some exciting and important studies to share later this year - from tracking the fate of pollinators to cracking protist biodiversity and tackling orphan crop genetics, as well as developing the tools with which the research community can better decode, understand, and share information about life on Earth.
One particular project to keep tabs on will be the Darwin Tree of Life, for which hundreds of new British and Irish eukaryotic genomes are set to be released to the community later this year.


If we’re to prosper, we must collectively cure this current malaise so that we might conserve and continue to benefit from biodiversity.

Nanopore sequencing using the Oxford Nanopore MinION has unlocked great potential for scientists looking to explore communities of organisms, from the human gut to rivers and oceans.
Dr Richard Leggett’s Technology Algorithms Group at the Earlham Institute was amongst the community of researchers that first helped to put this technology into practice, and has since developed a suite of tools - including NanoOK and MARTi - to help scientists analyse and understand the data generated.
This year, Leggett will be leading a brand new training course called “Nanopore metagenomics: from sample to analysis”, where users can learn from seasoned experts how to extract DNA from metagenomic samples. prepare libraries, carry out nanopore sequencing and analyse the data.
The registration deadline is 27 March, and it’s first come first served, so be sure to register soon if you don’t want to miss it!

Another area of expertise at the Earlham Institute is genome annotation, with the Swarbreck group in particular developing a range of software tools - such as Mikado and Portcullis - and pipelines to identify and build gene models.
Returning by popular demand, this 3-day course in May will help to provide scientists with an overview of eukaryotic genome annotation approaches. It covers advances in Next Generation Sequencing (NGS) technologies, transcriptome assembly, best practice guidance for building gene models utilising short and long read sequencing data or cross species proteins, and how to integrate and assess different gene models to go on to create a publication/release ready gene set.
The registration deadline is 17 April.
Our Head of Public Affairs, Saskia Hervey, has been working closely with colleagues such as Dr Nicola Patron to provide expert input during the UK’s careful deliberations over the potential of gene edited crops to improve agriculture, sustainability and the economy.
Later this month the government will introduce a Statutory Instrument (secondary legislation) aiming to make scientific research on gene edited crops easier, less expensive and less bureaucratic.
Although they will fall short of allowing commercial availability of gene edited plants, the measure is a precursor to a bigger piece of primary legislation that will pave the way for enabling a UK market in genetic engineered plants and animals.
According to a government resource online that provides more detail on what we can expect from the legislation, the UK plans “to pave the way to enable use of gene editing technologies, which can help better protect the environment.”

Author Dan Brown popularised the notion of the ‘golden ratio’ and its preeminence in living things in his popular book ‘The Da Vinci Code’, but the importance of mathematics in biology doesn’t stop there.
Dr Sam Martin in the Technology Algorithms Group was recently awarded a grant to apply novel mathematics to the study of phylogenetics - piecing together the evolutionary history of living things. He’ll be using algebraic techniques to help us understand phenomena such as horizontal gene transfer, showing us that the tree of life is more like an interconnected web.
Elsewhere in the institute, machine learning is increasingly being used to tackle a range of challenging topics, from understanding plant circadian rhythms to digesting the complex nature of our guts. Throughout the year we’ll be sharing the stories of this research, including the culmination of several exciting projects in collaboration with the Alan Turing Institute.
Last year we launched our ‘Barcoding the Broads’ public engagement programme in support of the Darwin Tree of Life Project, and throughout the spring and summer we’ll really be ramping up our offering - with workshops planned for a variety of nature groups and schools.
During the workshops, participants are introduced to easy-to-use techniques to collect, sample and analyse DNA from the environment using something known as DNA barcoding. This is an approach we’ve adopted from partners at the Cold Spring Harbour DNA Learning Centre.
If you’re interested in finding out more, please do get in touch.
We’ve also got exciting public engagement news to share, along with our collaborators and friends at the Darwin Tree of Life Consortium, which will hopefully give lots of you the opportunity to discover more about the project at one of the UK’s biggest and best science festivals.
Public Engagement Office, Dr Sam Rowe during one of our Barcoding workshops
Third time lucky, will this third instalment of the UK Conference of Bioinformatics and Computational Biology (UK CBCB) be the first attended in person? Only time will tell, but if the last two are anything to go by, this will be another fantastic opportunity for data-driven science enthusiasts to share work and explore the future of the field together.
Keep your eyes peeled for announcements of dates and registration in the coming months.
One of our New Year’s resolutions is to help more of you to achieve your research goals through the various facilities, platforms, and scientific services we offer.
Check out our new Digital Infrastructure pages to find out how our cloud computing resources can enable data-driven research in the life sciences. From virtual machines to expert advice, if you’re looking for the tools to get computational biology done, we’ve got you covered.
